n <- 1000 sim <- tibble( confounder = rbinom(n, 1, 0.5), p_exposure = case_when( confounder == 1 ~ 0.75, confounder == 0 ~ 0.25 ), exposure = rbinom(n, 1, p_exposure), outcome = confounder + rnorm(n) ) |> select(-p_exposure) simn <- 1000 sim <- tibble( confounder = rbinom(n, 1, 0.5), p_exposure = case_when( confounder == 1 ~ 0.75, confounder == 0 ~ 0.25 ), exposure = rbinom(n, 1, p_exposure), outcome = confounder + rnorm(n) ) |> select(-p_exposure) simn <- 1000 sim <- tibble( confounder = rbinom(n, 1, 0.5), p_exposure = case_when( confounder == 1 ~ 0.75, confounder == 0 ~ 0.25 ), exposure = rbinom(n, 1, p_exposure), outcome = confounder + rnorm(n) ) |> select(-p_exposure) simn <- 1000 sim <- tibble( confounder = rbinom(n, 1, 0.5), p_exposure = case_when( confounder == 1 ~ 0.75, confounder == 0 ~ 0.25 ), exposure = rbinom(n, 1, p_exposure), outcome = confounder + rnorm(n) ) |> select(-p_exposure) simn <- 1000 sim <- tibble( confounder = rbinom(n, 1, 0.5), p_exposure = case_when( confounder == 1 ~ 0.75, confounder == 0 ~ 0.25 ), exposure = rbinom(n, 1, p_exposure), outcome = confounder + rnorm(n) ) |> select(-p_exposure) sim
Causal Inference with group_by and summarise
Lucy D’Agostino McGowan
Wake Forest University
Observational Studies
Goal: To answer a research question
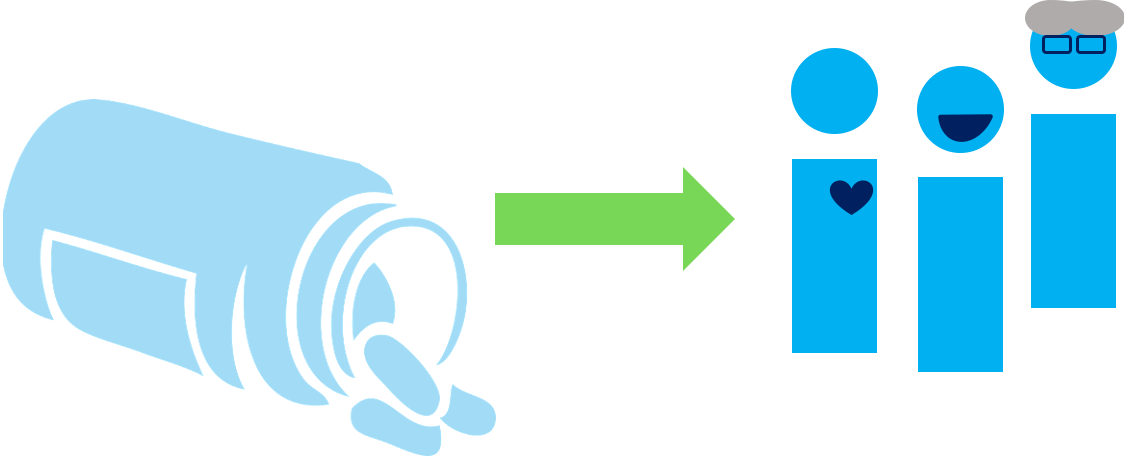
Observational Studies
Goal: To answer a research question
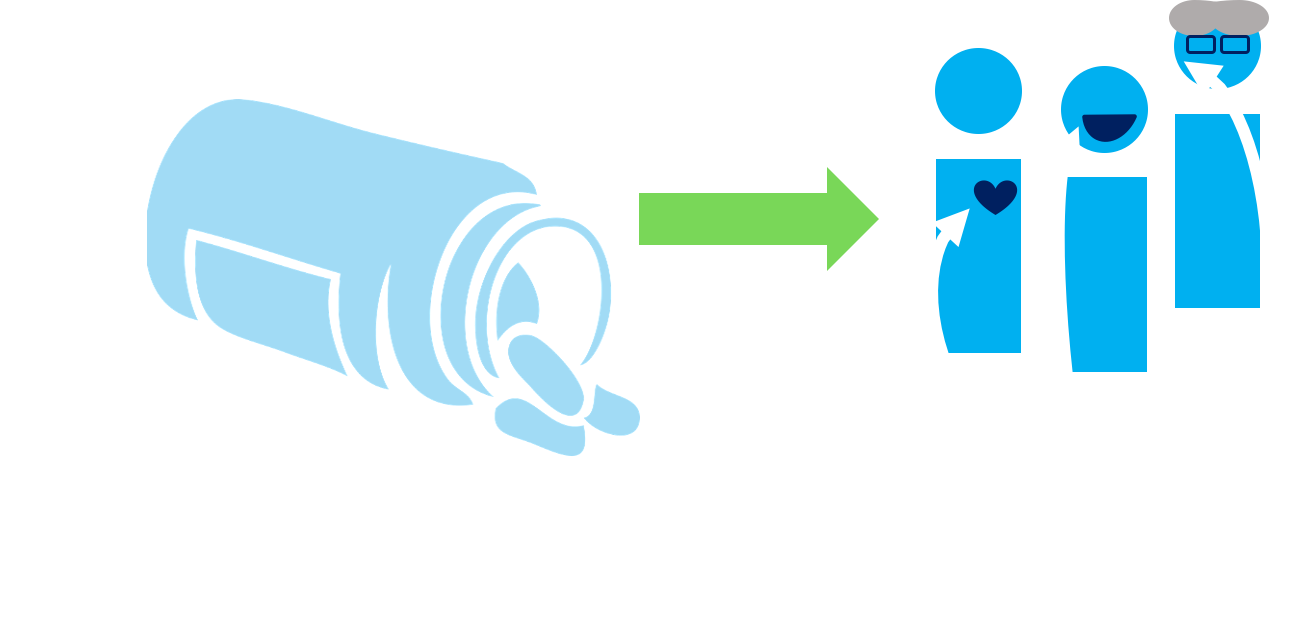
Observational Studies
Randomized Controlled Trial
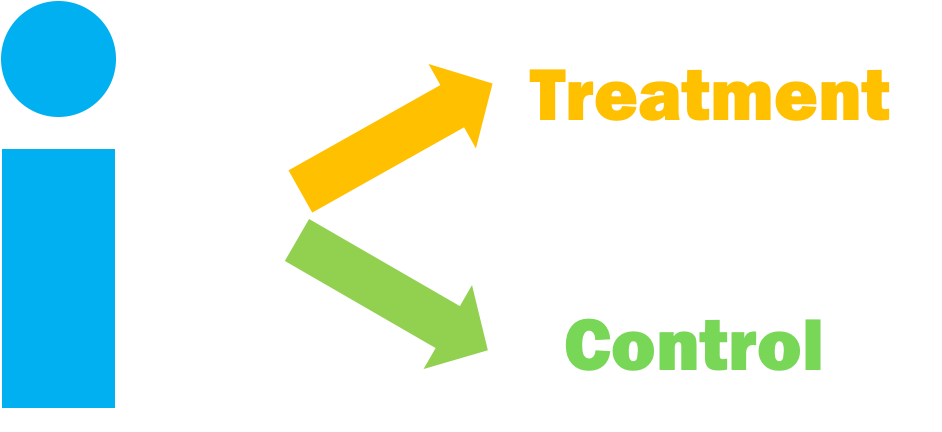
Observational Studies
Randomized Controlled Trial
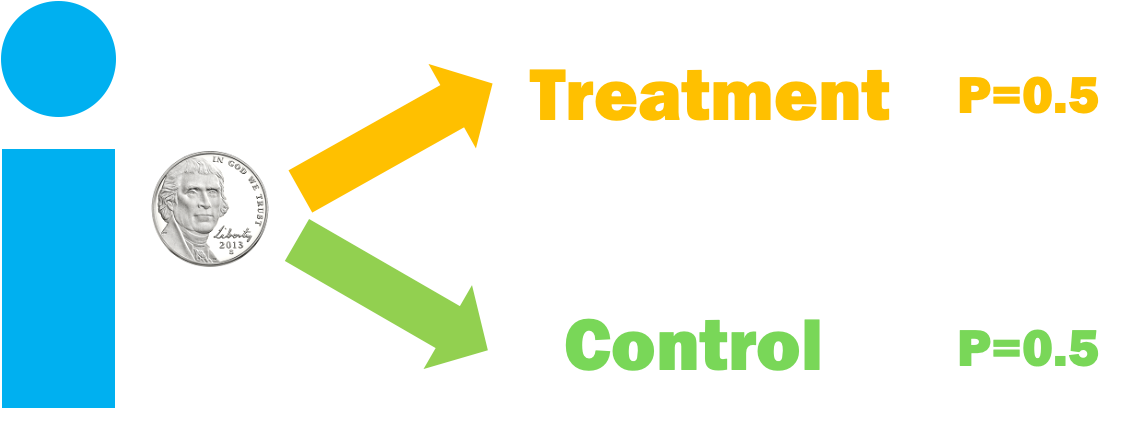
Observational Studies

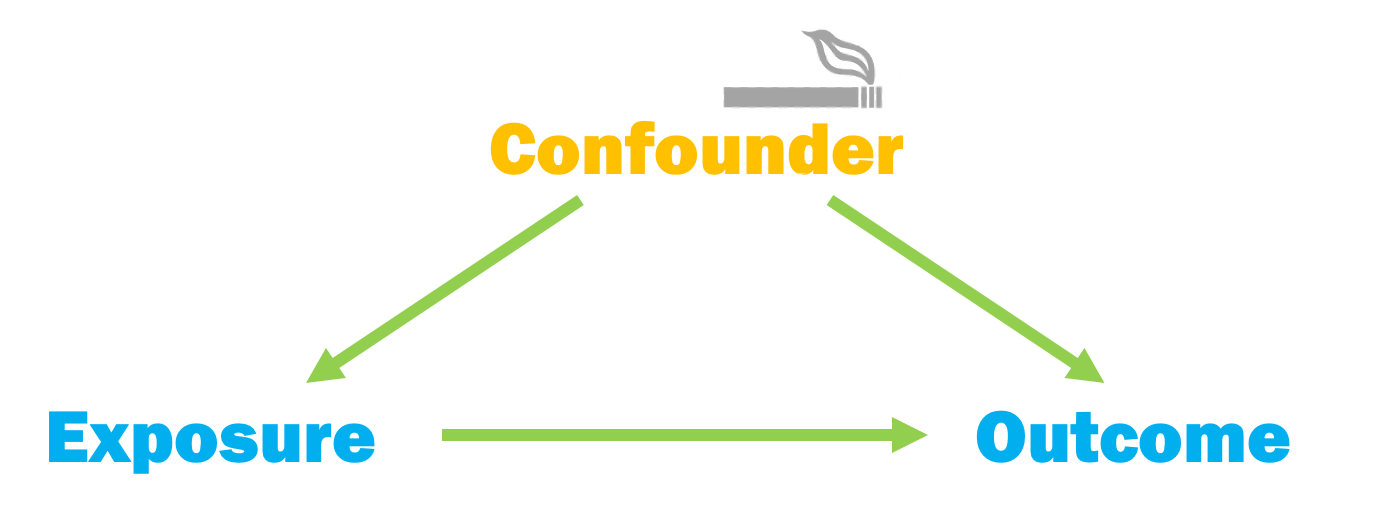
Confounding

Confounding
One binary confounder
Simulation
Simulation
# A tibble: 1,000 × 3
confounder exposure outcome
<int> <int> <dbl>
1 0 0 1.13
2 0 0 1.11
3 1 1 0.129
4 1 0 1.21
5 0 0 0.0694
6 1 1 -0.663
7 1 1 1.81
8 1 1 -0.912
9 1 0 -0.247
10 0 0 0.998
# ℹ 990 more rowsSimulation
Simulation
# A tibble: 2 × 2
exposure avg_y
<int> <dbl>
1 0 0.269
2 1 0.676Simulation
Your Turn 1 (03-ci-with-group-by-and-summarise-exercises.qmd)
Group the dataset by confounder and exposure
Calculate the mean of the outcome for the groups
03:00 Your Turn 1
# A tibble: 4 × 3
# Groups: confounder [2]
confounder exposure avg_y
<int> <int> <dbl>
1 0 0 -0.00907
2 0 1 -0.0166
3 1 0 1.09
4 1 1 0.936 Your Turn 1
sim |> group_by(confounder, exposure) |> summarise(avg_y = mean(outcome)) |> pivot_wider( names_from = exposure, values_from = avg_y, names_prefix = "x_" ) |> summarise(estimate = x_1 - x_0) |> # note: we would need to weight this # if the confounder groups were not equal sized summarise(estimate = mean(estimate))sim |> group_by(confounder, exposure) |> summarise(avg_y = mean(outcome)) |> pivot_wider( names_from = exposure, values_from = avg_y, names_prefix = "x_" ) |> summarise(estimate = x_1 - x_0) |> # note: we would need to weight this # if the confounder groups were not equal sized summarise(estimate = mean(estimate))
# A tibble: 1 × 1
estimate
<dbl>
1 -0.0794🎉
Two binary confounders
Simulation
n <- 1000 sim2 <- tibble( confounder_1 = rbinom(n, 1, 0.5), confounder_2 = rbinom(n, 1, 0.5), p_exposure = case_when( confounder_1 == 1 & confounder_2 == 1 ~ 0.75, confounder_1 == 0 & confounder_2 == 1 ~ 0.9, confounder_1 == 1 & confounder_2 == 0 ~ 0.2, confounder_1 == 0 & confounder_2 == 0 ~ 0.1, ), exposure = rbinom(n, 1, p_exposure), outcome = confounder_1 + confounder_2 + rnorm(n) ) |> select(-p_exposure) sim2n <- 1000 sim2 <- tibble( confounder_1 = rbinom(n, 1, 0.5), confounder_2 = rbinom(n, 1, 0.5), p_exposure = case_when( confounder_1 == 1 & confounder_2 == 1 ~ 0.75, confounder_1 == 0 & confounder_2 == 1 ~ 0.9, confounder_1 == 1 & confounder_2 == 0 ~ 0.2, confounder_1 == 0 & confounder_2 == 0 ~ 0.1, ), exposure = rbinom(n, 1, p_exposure), outcome = confounder_1 + confounder_2 + rnorm(n) ) |> select(-p_exposure) sim2
Simulation
# A tibble: 1,000 × 4
confounder_1 confounder_2 exposure outcome
<int> <int> <int> <dbl>
1 0 0 0 0.521
2 1 0 0 1.38
3 0 0 0 -0.624
4 0 1 1 0.427
5 1 0 1 1.31
6 0 0 0 -0.707
7 1 1 1 2.52
8 1 0 0 1.45
9 0 0 0 -0.505
10 0 1 1 0.793
# ℹ 990 more rowsSimulation
Your Turn 2
Group the dataset by the confounders and exposure
Calculate the mean of the outcome for the groups
02:00 Your Turn 2
sim2 |> group_by(confounder_1, confounder_2, exposure) |> summarise(avg_y = mean(outcome)) |> pivot_wider( names_from = exposure, values_from = avg_y, names_prefix = "x_" ) |> summarise(estimate = x_1 - x_0, .groups = "drop") |> summarise(estimate = mean(estimate))sim2 |> group_by(confounder_1, confounder_2, exposure) |> summarise(avg_y = mean(outcome)) |> pivot_wider( names_from = exposure, values_from = avg_y, names_prefix = "x_" ) |> summarise(estimate = x_1 - x_0, .groups = "drop") |> summarise(estimate = mean(estimate))
# A tibble: 1 × 1
estimate
<dbl>
1 -0.0731Simulation
n <- 100000 big_sim2 <- tibble( confounder_1 = rbinom(n, 1, 0.5), confounder_2 = rbinom(n, 1, 0.5), p_exposure = case_when( confounder_1 == 1 & confounder_2 == 1 ~ 0.75, confounder_1 == 0 & confounder_2 == 1 ~ 0.9, confounder_1 == 1 & confounder_2 == 0 ~ 0.2, confounder_1 == 0 & confounder_2 == 0 ~ 0.1, ), exposure = rbinom(n, 1, p_exposure), outcome = confounder_1 + confounder_2 + rnorm(n) ) |> select(-p_exposure) big_sim2n <- 100000 big_sim2 <- tibble( confounder_1 = rbinom(n, 1, 0.5), confounder_2 = rbinom(n, 1, 0.5), p_exposure = case_when( confounder_1 == 1 & confounder_2 == 1 ~ 0.75, confounder_1 == 0 & confounder_2 == 1 ~ 0.9, confounder_1 == 1 & confounder_2 == 0 ~ 0.2, confounder_1 == 0 & confounder_2 == 0 ~ 0.1, ), exposure = rbinom(n, 1, p_exposure), outcome = confounder_1 + confounder_2 + rnorm(n) ) |> select(-p_exposure) big_sim2
Simulation
# A tibble: 100,000 × 4
confounder_1 confounder_2 exposure outcome
<int> <int> <int> <dbl>
1 1 1 1 2.35
2 1 1 0 3.71
3 0 0 0 2.08
4 0 1 1 0.516
5 0 0 0 -0.166
6 1 1 1 1.58
7 0 0 0 0.472
8 1 0 0 3.22
9 0 1 1 0.929
10 0 1 1 1.41
# ℹ 99,990 more rowsSimulation
Simulation
big_sim2 |> group_by(confounder_1, confounder_2, exposure) |> summarise(avg_y = mean(outcome)) |> pivot_wider( names_from = exposure, values_from = avg_y, names_prefix = "x_" ) |> summarise(estimate = x_1 - x_0, .groups = "drop") |> summarise(estimate = mean(estimate))big_sim2 |> group_by(confounder_1, confounder_2, exposure) |> summarise(avg_y = mean(outcome)) |> pivot_wider( names_from = exposure, values_from = avg_y, names_prefix = "x_" ) |> summarise(estimate = x_1 - x_0, .groups = "drop") |> summarise(estimate = mean(estimate))
# A tibble: 1 × 1
estimate
<dbl>
1 0.0187Continuous confounder?
Simulation
n <- 10000 sim3 <- tibble( confounder = rnorm(n), p_exposure = exp(confounder) / (1 + exp(confounder)), exposure = rbinom(n, 1, p_exposure), outcome = confounder + rnorm(n) ) |> select(-p_exposure) sim3n <- 10000 sim3 <- tibble( confounder = rnorm(n), p_exposure = exp(confounder) / (1 + exp(confounder)), exposure = rbinom(n, 1, p_exposure), outcome = confounder + rnorm(n) ) |> select(-p_exposure) sim3
Simulation
# A tibble: 10,000 × 3
confounder exposure outcome
<dbl> <int> <dbl>
1 -0.167 0 -0.560
2 0.252 1 0.628
3 -0.321 1 -0.608
4 0.621 0 1.58
5 -0.619 1 0.358
6 -0.897 0 -1.95
7 -2.01 0 -2.50
8 0.296 0 -1.10
9 -0.504 1 -0.316
10 -0.536 1 1.12
# ℹ 9,990 more rowsSimulation
Your Turn 3
Use ntile() from dplyr to calculate a binned version of confounder called confounder_q. We’ll create a variable with 5 bins.
Group the dataset by the binned variable you just created and exposure
Calculate the mean of the outcome for the groups
03:00 Your Turn 3
sim3 |> mutate(confounder_q = ntile(confounder, 5)) |> group_by(confounder_q, exposure) |> summarise(avg_y = mean(outcome)) |> pivot_wider( names_from = exposure, values_from = avg_y, names_prefix = "x_" ) |> summarise(estimate = x_1 - x_0) |> summarise(estimate = mean(estimate))sim3 |> mutate(confounder_q = ntile(confounder, 5)) |> group_by(confounder_q, exposure) |> summarise(avg_y = mean(outcome)) |> pivot_wider( names_from = exposure, values_from = avg_y, names_prefix = "x_" ) |> summarise(estimate = x_1 - x_0) |> summarise(estimate = mean(estimate))sim3 |> mutate(confounder_q = ntile(confounder, 5)) |> group_by(confounder_q, exposure) |> summarise(avg_y = mean(outcome)) |> pivot_wider( names_from = exposure, values_from = avg_y, names_prefix = "x_" ) |> summarise(estimate = x_1 - x_0) |> summarise(estimate = mean(estimate))
# A tibble: 1 × 1
estimate
<dbl>
1 0.0728What if we could come up with a summary score of all confounders?
Causal Inference with group_by and summarise Lucy D’Agostino McGowan Wake Forest University