Continuous exposures with propensity scores
Malcolm Barrett
Stanford University
Warning! Propensity score weights are sensitive to positivity violations for continuous exposures.
The story so far
Propensity score weighting
- Fit a propensity model predicting exposure
x,x + zwhere z is all covariates - Calculate weights
- Fit an outcome model estimating the effect of
xonyweighted by the propensity score
Continous exposures
- Use a model like
lm(x ~ z)for the propensity score model. - Use
wt_ate()with.fittedand.sigma; transforms usingdnorm()to get on probability-like scale. - Apply the weights to the outcome model as normal!
Alternative: quantile binning
- Bin the continuous exposure into quantiles and use categorical regression like a multinomial model to calculate probabilities.
- Calculate the weights where the propensity score is the probability you fall into the quantile you actually fell into. Same as the binary ATE!
- Same workflow for the outcome model
1. Fit a model for exposure ~ confounders
2. Calculate the weights with wt_ate()
Does change in smoking intensity (smkintensity82_71) affect weight gain among lighter smokers?
1. Fit a model for exposure ~ confounders
nhefs_model <- lm( smkintensity82_71 ~ sex + race + age + I(age^2) + education + smokeintensity + I(smokeintensity^2) + smokeyrs + I(smokeyrs^2) + exercise + active + wt71 + I(wt71^2), data = nhefs_light_smokers )nhefs_model <- lm( smkintensity82_71 ~ sex + race + age + I(age^2) + education + smokeintensity + I(smokeintensity^2) + smokeyrs + I(smokeyrs^2) + exercise + active + wt71 + I(wt71^2), data = nhefs_light_smokers )nhefs_model <- lm( smkintensity82_71 ~ sex + race + age + I(age^2) + education + smokeintensity + I(smokeintensity^2) + smokeyrs + I(smokeyrs^2) + exercise + active + wt71 + I(wt71^2), data = nhefs_light_smokers )
2. Calculate the weights with wt_ate()
2. Calculate the weights with wt_ate()
# A tibble: 1,162 × 74
seqn qsmk death yrdth modth dadth sbp dbp sex
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <fct>
1 235 0 0 NA NA NA 123 80 0
2 244 0 0 NA NA NA 115 75 1
3 245 0 1 85 2 14 148 78 0
4 252 0 0 NA NA NA 118 77 0
5 257 0 0 NA NA NA 141 83 1
6 262 0 0 NA NA NA 132 69 1
7 266 0 0 NA NA NA 100 53 1
8 419 0 1 84 10 13 163 79 0
9 420 0 1 86 10 17 184 106 0
10 434 0 0 NA NA NA 127 80 1
# ℹ 1,152 more rows
# ℹ 65 more variables: age <dbl>, race <fct>, income <dbl>,
# marital <dbl>, school <dbl>, education <fct>, …Do posted wait times at 8 am affect actual wait times at 9 am?
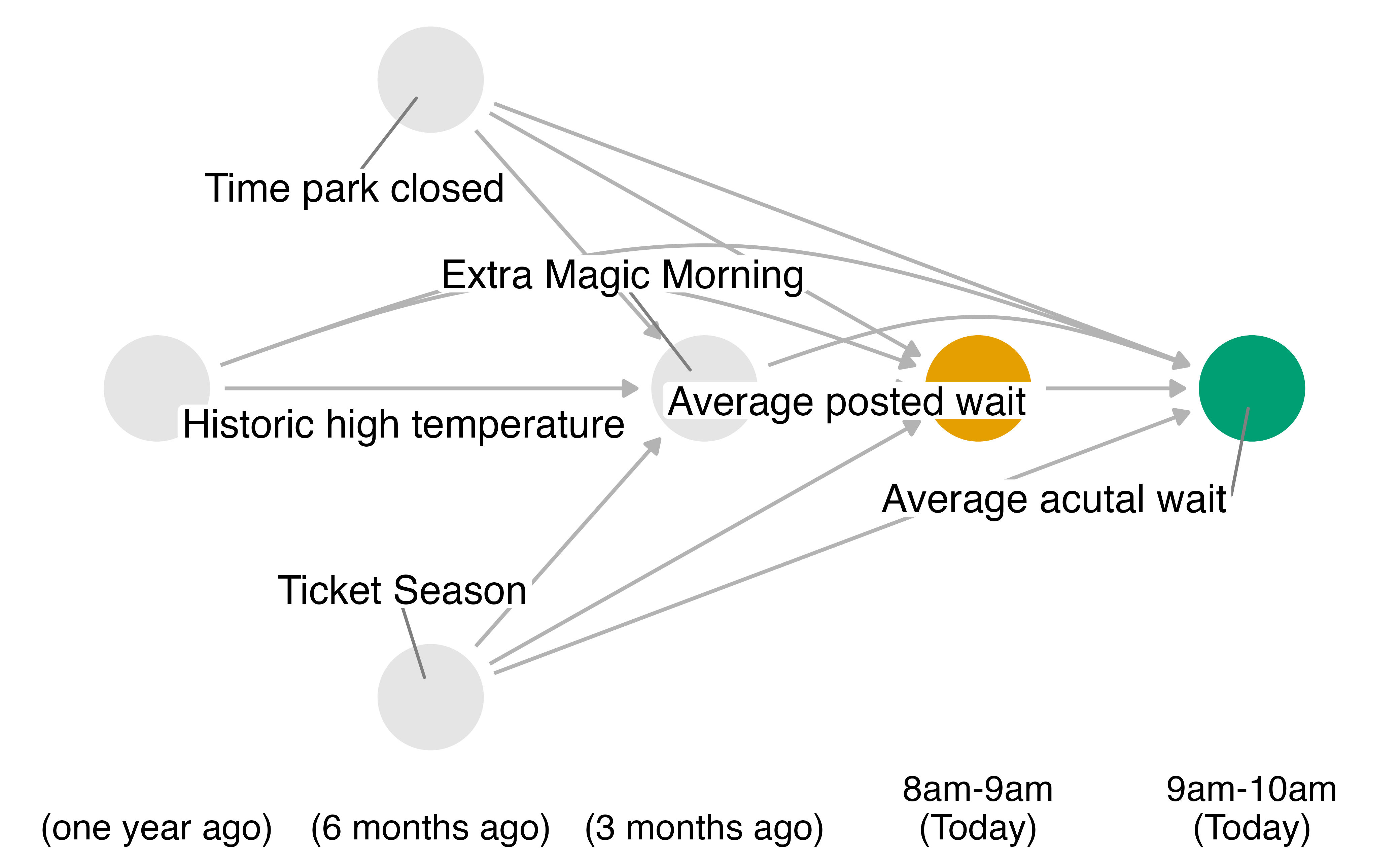
Your Turn 1
Fit a model using lm() with wait_minutes_posted_avg as the outcome and the confounders identified in the DAG.
Use augment() to add model predictions to the data frame
In wt_ate(), calculate the weights using wait_minutes_posted_avg, .fitted, and .sigma
05:00 Your Turn 1
Your Turn 1
Stabilizing extreme weights
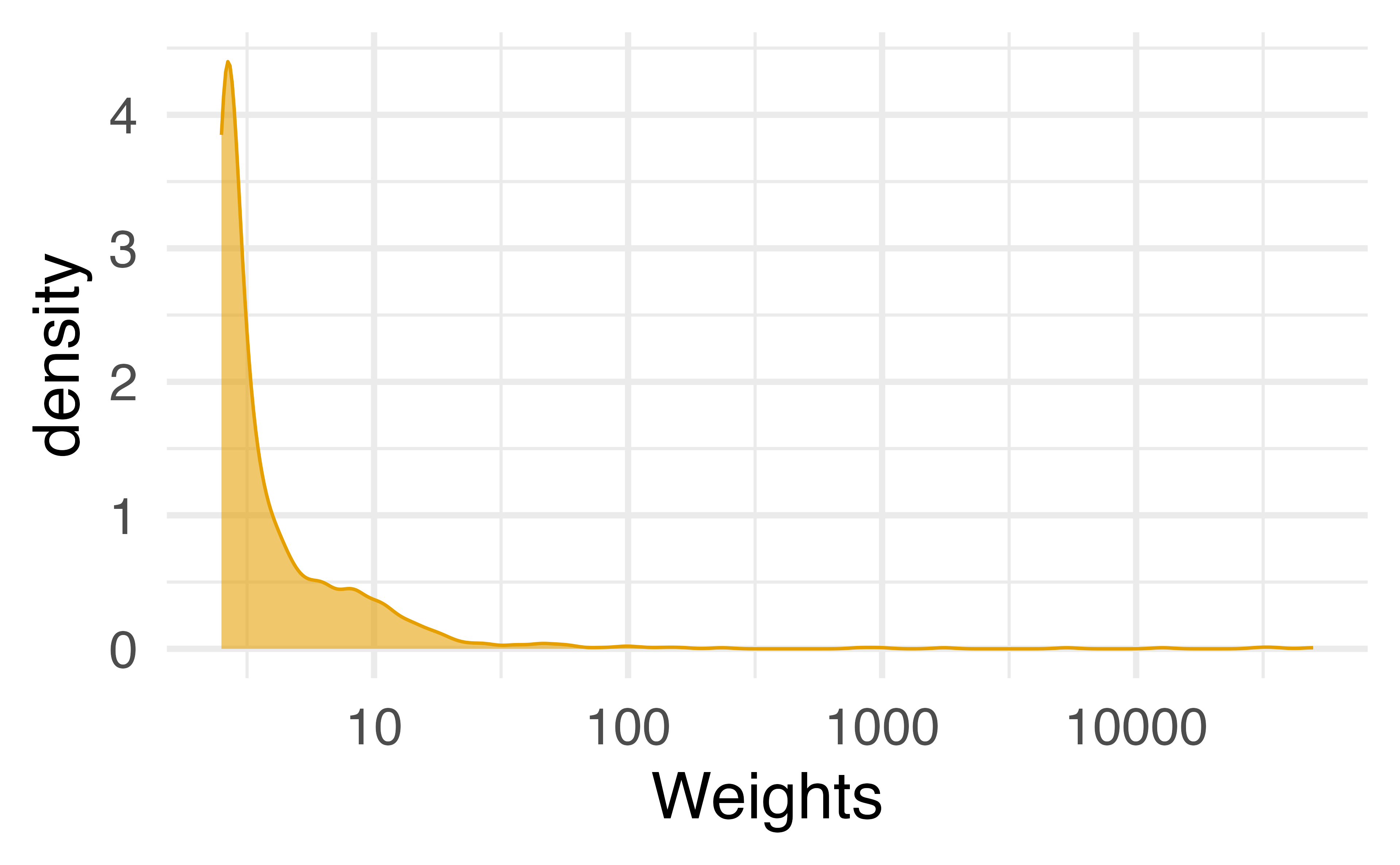
Stabilizing extreme weights
- Fit an intercept-only model (e.g.
lm(x ~ 1)) or use mean and SD ofx - Calculate weights from this model.
- Divide these weights by the propensity score weights.
wt_ate(.., stabilize = TRUE)does this all!
Calculate stabilized weights
nhefs_swts <- nhefs_model |> augment(data = nhefs_light_smokers) |> mutate(swts = wt_ate( smkintensity82_71, .fitted, .sigma = .sigma, stabilize = TRUE ))nhefs_swts <- nhefs_model |> augment(data = nhefs_light_smokers) |> mutate(swts = wt_ate( smkintensity82_71, .fitted, .sigma = .sigma, stabilize = TRUE ))
Stabilizing extreme weights
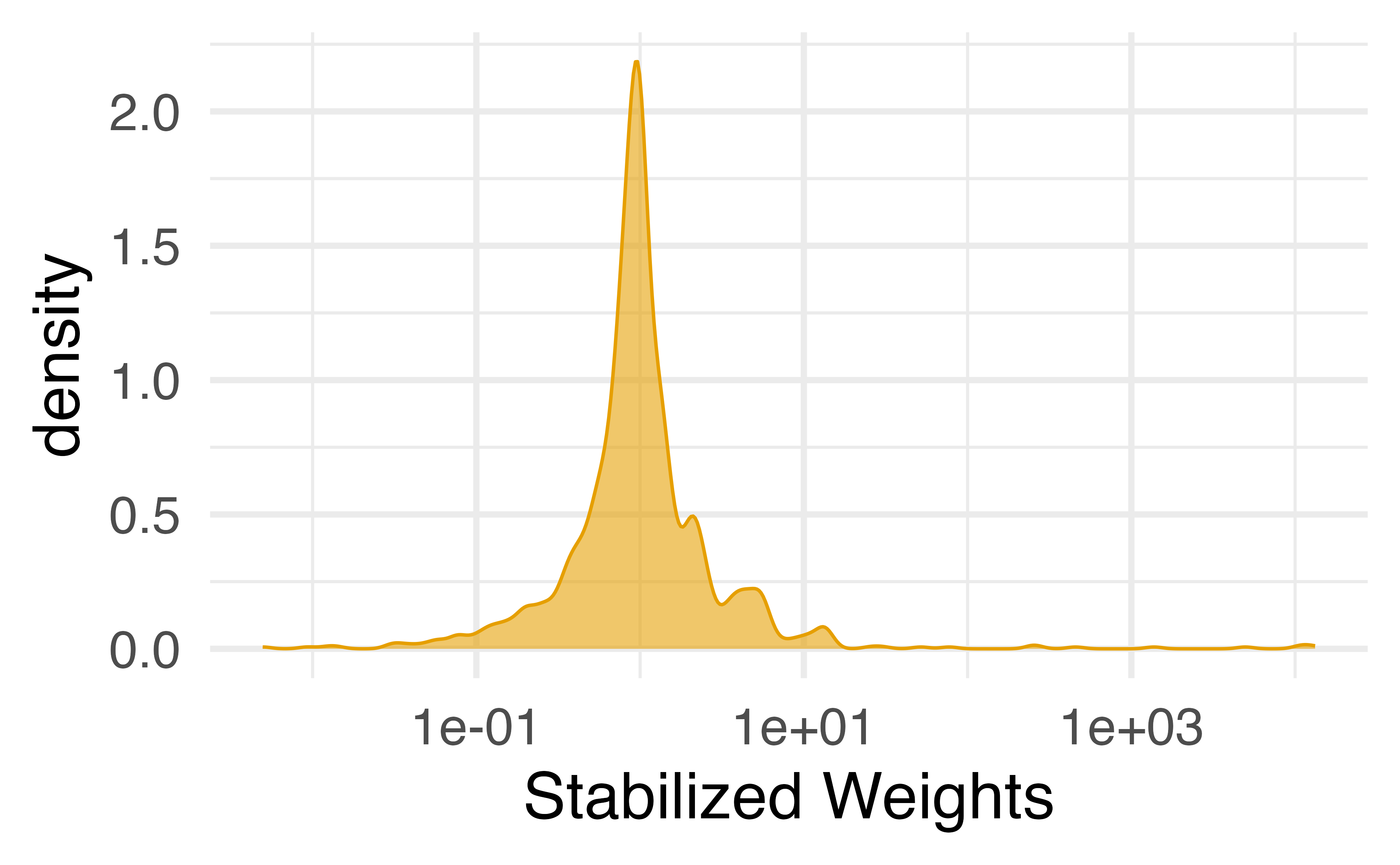
Your Turn 2
Re-fit the above using stabilized weights
03:00 Your Turn 2
Fitting the outcome model
- Use the stabilized weights in the outcome model. Nothing new here!
lm( wt82_71 ~ smkintensity82_71, weights = swts, data = nhefs_swts ) |> tidy() |> filter(term == "smkintensity82_71") |> mutate(estimate = estimate * -10)lm( wt82_71 ~ smkintensity82_71, weights = swts, data = nhefs_swts ) |> tidy() |> filter(term == "smkintensity82_71") |> mutate(estimate = estimate * -10)
# A tibble: 1 × 5
term estimate std.error statistic p.value
<chr> <dbl> <dbl> <dbl> <dbl>
1 smkintensity82_71 2.04 0.0335 -6.08 1.62e-9Your Turn 3
Estimate the relationship between posted wait times and actual wait times using the stabilized weights we just created.
03:00 Your Turn 3
lm(
wait_minutes_actual_avg ~ wait_minutes_posted_avg,
weights = swts,
data = wait_times_swts
) |>
tidy() |>
filter(term == "wait_minutes_posted_avg") |>
mutate(estimate = estimate * 10)# A tibble: 1 × 5
term estimate std.error statistic p.value
<chr> <dbl> <dbl> <dbl> <dbl>
1 wait_minutes_posted_… 2.40 0.0655 3.66 4.48e-4Diagnosing issues
- Extreme weights even after stabilization
- Bootstrap: non-normal distribution
- Bootstrap: estimate different from original model
More info
https://github.com/LucyMcGowan/writing-positivity-continous-ps
Continuous exposures with propensity scores Malcolm Barrett Stanford University